Virus Explorer
Click and drag or use the buttons below to move the model.
Use the tab to focus one of the buttons above, and then space to start the rotation.
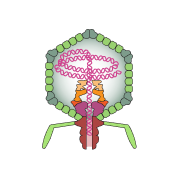
The proteins that make up the icosahedral capsid and tail structure are shown.
Components on the virus are selectively represented in the 3D model for educational purposes.
A. Capsid proteins; B. DNA genome; C. Core proteins; D. Portal protein; E. Tail fiber; F. Tail proteins; G. Tail; H. Head
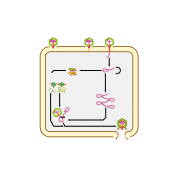
This diagram shows how the T7 virus replicates, or makes copies of itself. First, the tail fiber proteins on the virus’s surface bind to specific receptors on the surface of a bacterial host cell (A). The virus makes an opening in the cell membrane (B), through which it injects its dsDNA genome into the cell’s cytosol (C).
With the help of the host cell’s machinery, the virus’s DNA is transcribed into mRNAs (E), which are translated into proteins by ribosomes in the cytosol (F). Some of these proteins are used to build parts of the new viruses (G). The cell also helps the virus make one long DNA strand containing many repeating copies of the virus’s genome (D). This strand is cleaved, or cut, into separate genomes that are packaged into new viruses (H).
Once the new viruses become fully formed through a series of changes called virus maturation (I), they leave the cell. The viruses are released through a process called cell lysis (J), which kills the host cell.
Phages like T7 usually cause cell lysis of their hosts (known as a lytic infection). Other types of phages insert their genome into the host cell’s genome (known as a lysogenic infection). The inserted viral DNA is replicated by the host cell, and descendants of the host cell will also contain the viral DNA. Although new viruses are not actively produced, viral production may resume later depending on cellular conditions.
T7 virus
- Part of the Autographiviridae family
- ~60-nm naked icosahedral capsid with an attached tail structure
- Linear dsDNA genome of ~40,000 bp
- Infects bacteria, specifically Escherichia coli
T7 viruses are phages, which are viruses that infect bacteria. Most phages are easily distinguished from other types of viruses by their unique head-and-tail structure.
T7 viruses are used extensively in modern molecular biology research. Scientists can manipulate the genomes and proteins of these viruses to study gene expression, protein interactions, and more.