Virus Explorer
Click and drag or use the buttons below to move the model.
Use the tab to focus one of the buttons above, and then space to start the rotation.
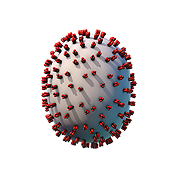
The viral envelope (gray) and proteins embedded in the envelope (red) are shown.
Components on the virus are selectively represented in the 3D model for educational purposes.
A. Hemagglutinin glycoprotein; B. Neuraminidase glycoprotein; C. Ion channel; D. Polymerase complex; E. Nucleocapsid protein; F. Nuclear export protein; G. RNA genome; H. Matrix protein; I. Lipid envelope
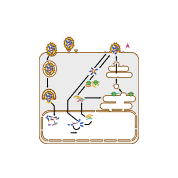
This diagram shows how the influenza A virus replicates, or makes copies of itself. Glycoproteins on the surface of the influenza A virus bind to specific receptors on the host cell’s surface (A). This triggers a process called endocytosis (B), which brings the virus into the cell in an enclosed structure (vesicle) called an endosome. Acidic substances build up inside the endosome, lowering the pH and triggering the endosome’s membrane to fuse with the virus’s envelope. This releases the virus’s (–)RNA genome segments and viral polymerase into the cell’s cytosol (C).
The viral genome enters the cell’s nucleus, where the viral polymerase transcribes the virus’s (–)RNA genome segments into complementary (+)RNA templates (D). These templates are used to make copies of all the virus’s (–)RNA genome segments (E).
The virus’s (–)RNA segments are also transcribed into mRNAs (F), which are translated into proteins by ribosomes in the cytosol and on the endoplasmic reticulum (ER) (G). Influenza virus glycoproteins go through protein processing in the ER-Golgi network (H), then are transported to the cell membrane.
Viral genomes and proteins are assembled into new viruses (I), which leave the cell through a process called budding (J). This process surrounds the virus in a piece of the cell membrane containing viral proteins, which becomes the virus’s envelope.
Influenza A virus
- Part of the Orthomyxoviridae family, which also includes other types of influenza viruses
- ~100-nm enveloped particles with a spherical or oblong capsid
- Segmented (–)ssRNA genome of ~13,500 bp, consisting of eight segments
- Infects birds and mammals including humans and pigs
- Vaccine produced for new seasonal subtypes each year
There are four species of influenza viruses (A, B, C, D), which vary in the severity of symptoms they cause and in their host range. Influenza A and B viruses cause outbreaks of influenza (also known as the flu) almost every winter worldwide. The main types of influenza viruses are further divided into different varieties called subtypes.
Influenza A viruses have a segmented genome, which means that their genome has multiple separate sections that are not joined to each other. When two or more influenza viruses infect a single cell, their genome segments can rearrange to produce a new combination of segments, giving rise to a new virus subtype. Every year, different subtypes of influenza A viruses infect humans, causing seasonal outbreaks of flu. Sometimes, dangerous subtypes emerge, such as the 2009 H1N1 pandemic influenza, also called swine flu. This subtype had a combination of genome segments from human, avian, and swine influenza viruses.